S2: Anatomical Cord Reference
Step Code: S2_anat_cordref
Depends on: S1 (Input Verification)
Required by: S3-S11 (all downstream steps)
Purpose
S2 establishes the anatomical cord reference that anchors all subsequent fMRI processing. The spinal cord presents unique challenges compared to brain imaging: its small cross-sectional area (~50-80 mm²), susceptibility to physiological motion, and variable vertebral anatomy across subjects. This step addresses these challenges by:
- Creating a standardized anatomical reference for downstream registration
- Segmenting the spinal cord for region-of-interest analyses
- Labeling vertebral levels for spatial normalization
- Registering to the PAM50 template for group-level analyses
Algorithm Overview
The S2 step consists of four sequential substeps:
| Substep | Name | Description |
|---|---|---|
| S2.1 | Discovery + Crop | Select anatomy, standardize, discover cord, crop |
| S2.2 | Segmentation + Labels | Cord segmentation, TotalSpineSeg for vertebrae/discs/canal |
| S2.3 | Rootlets | Dorsal nerve rootlet detection (conditional) |
| S2.4 | Registration | PAM50 template registration with dual-path strategy |
S2.1: Discovery and Cropping
Rationale
Spinal cord MRI often includes significant non-cord tissue (e.g., brainstem, shoulders) that can interfere with segmentation and registration algorithms. The discovery phase identifies the cord location and creates a cropped working volume.
Algorithm
- Anatomy Selection - Prefer T2w (better cord-CSF contrast), fallback to T1w
- Standardization - Reorient to RPI via
sct_image -setorient RPI - Discovery Segmentation -
sct_deepseg -task seg_sc_contrast_agnostic - FOV Validation - Reject if cord coverage < 20 slices
- Cord-Focused Cropping -
sct_crop_image -m <crop_mask>
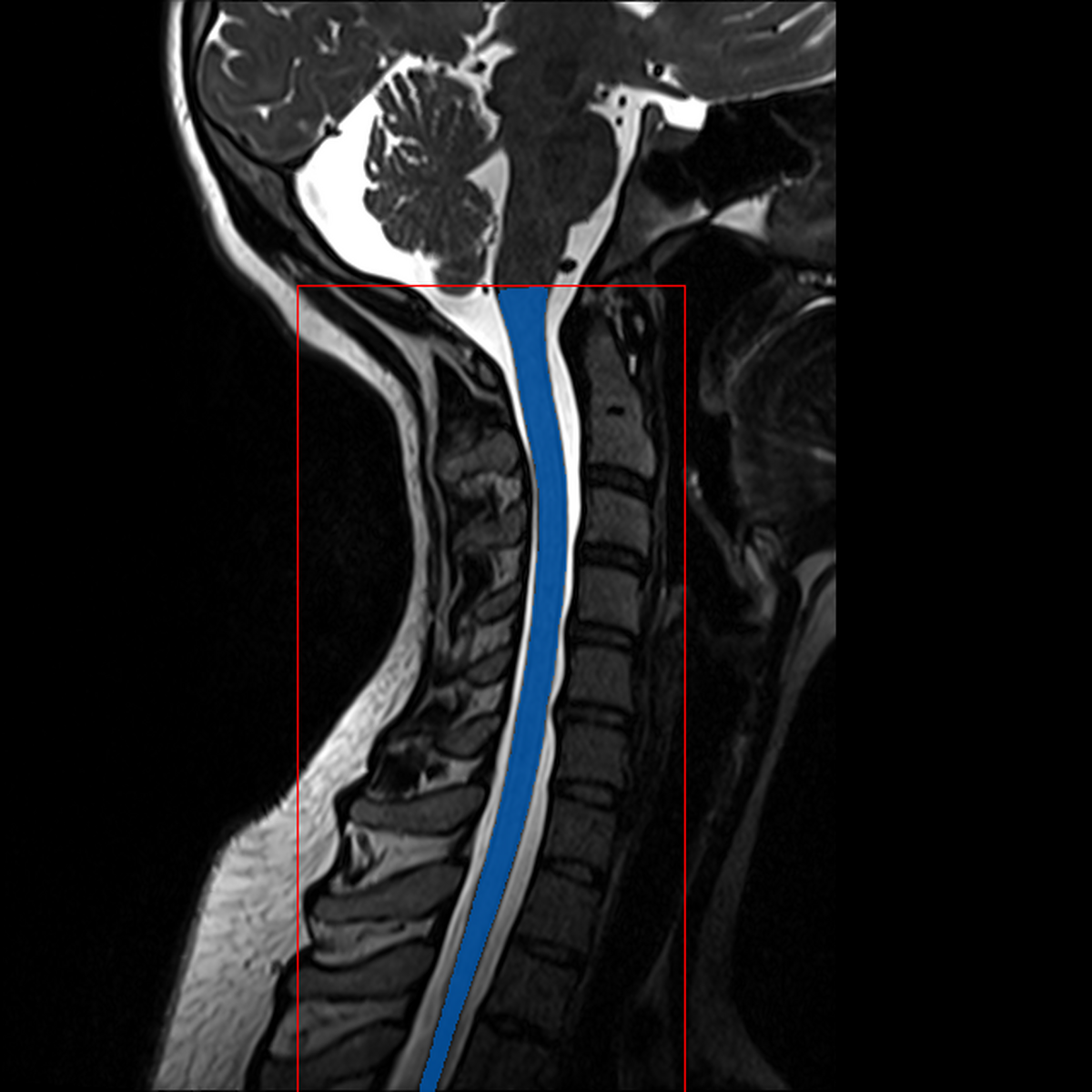
QC: Crop Box Sagittal
What to look for:
- ✅ Blue contour (cord mask) follows the spinal cord
- ✅ Red contour (crop box) encompasses the full cord region
- ❌ FAIL: Crop box extends into brain or misses cord
S2.2: Segmentation and Labeling
Cord Segmentation Methods
| Method | Command | Best For |
|---|---|---|
contrast_agnostic (default) | sct_deepseg -task seg_sc_contrast_agnostic | CSA consistency |
totalspineseg | Extract from TSS output | All-in-one |
TotalSpineSeg Integration
TotalSpineSeg provides automatic instance segmentation:
- Spinal cord (label 1)
- Spinal canal (label 2)
- Vertebrae (labels 11-50: C1 through Sacrum)
- Intervertebral discs (labels 63-100: C2/C3 through L5/S)
Command: sct_deepseg -task totalspineseg
Metrics Computed
- Cord volume (mm³), length (mm)
- CSA: mean, min, max (mm²)
- Vertebrae/discs detected
- Canal volume
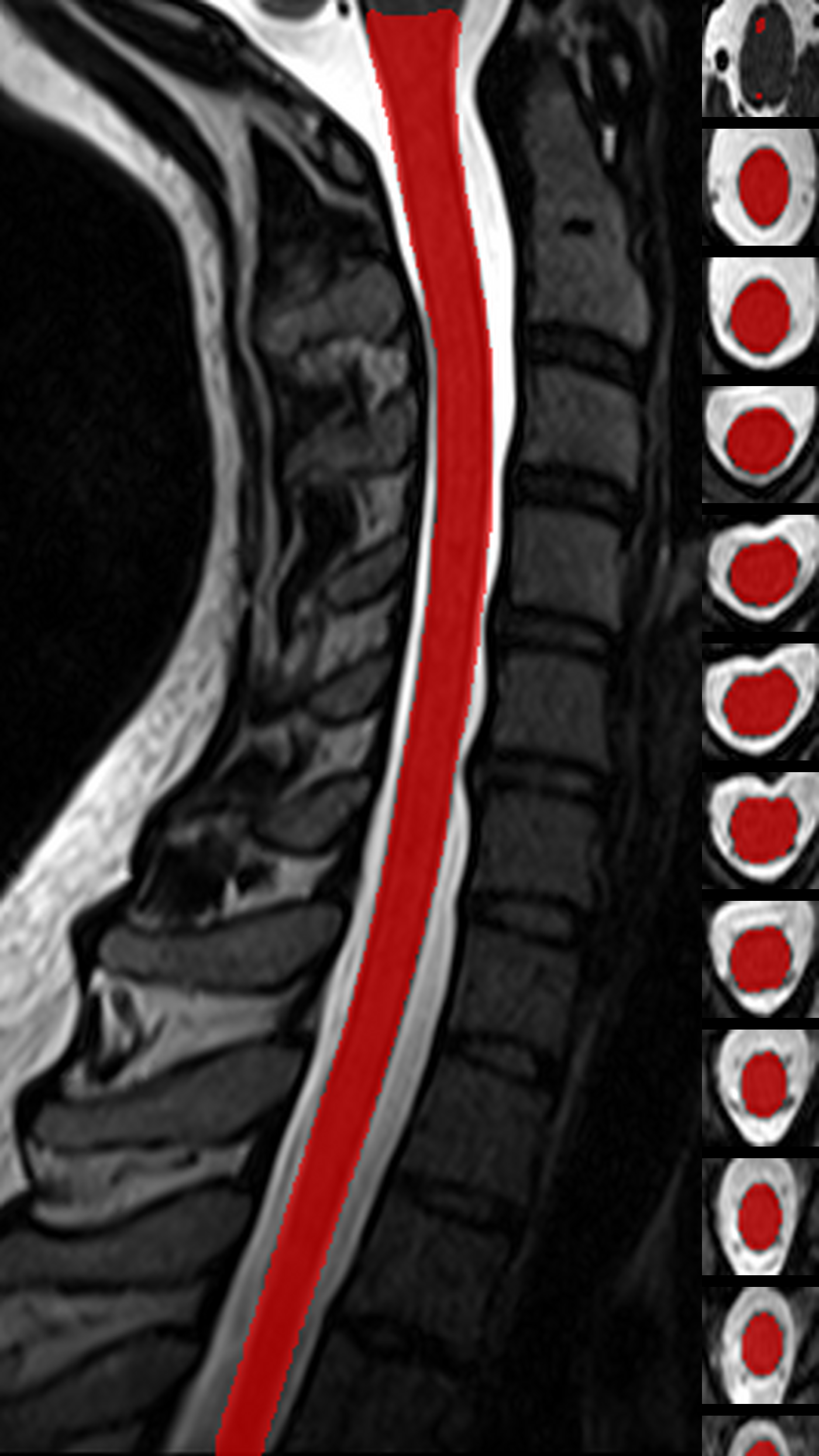
QC: Cord Segmentation Montage
What to look for:
- ✅ Cord mask is continuous across all slices
- ✅ Mask is centered on the cord
- ❌ FAIL: Large gaps or leakage into CSF
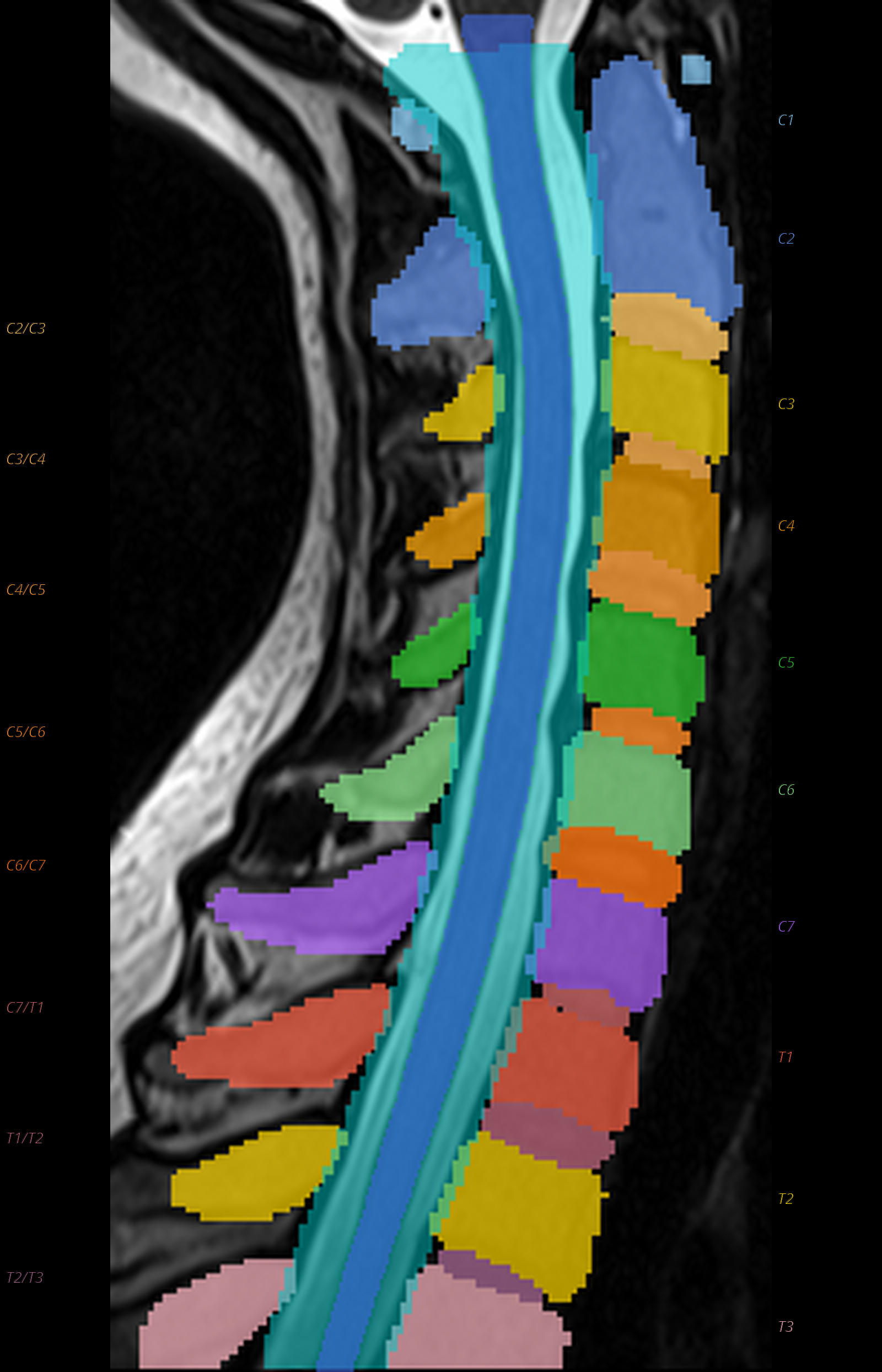
QC: TotalSpineSeg Montage
Sagittal view showing colored vertebrae, discs, cord (blue), canal (cyan), with anatomical labels.
What to look for:
- ✅ Vertebrae are correctly ordered (no label jumps)
- ✅ Disc labels correspond to adjacent vertebrae
- ❌ FAIL: Missing vertebrae or shifted labels
S2.3: Rootlets Detection
Eligibility
- T2w image available
- Sufficient cord coverage
- Policy-enabled
Command: sct_deepseg -task seg_spinal_rootlets
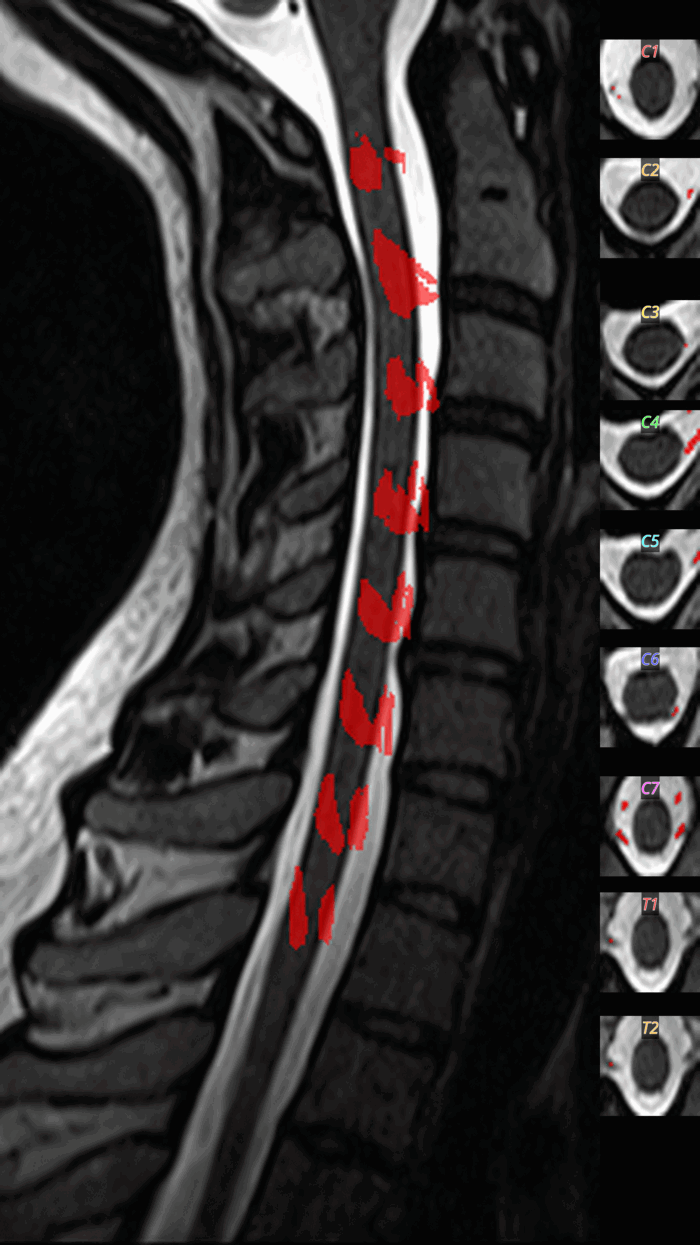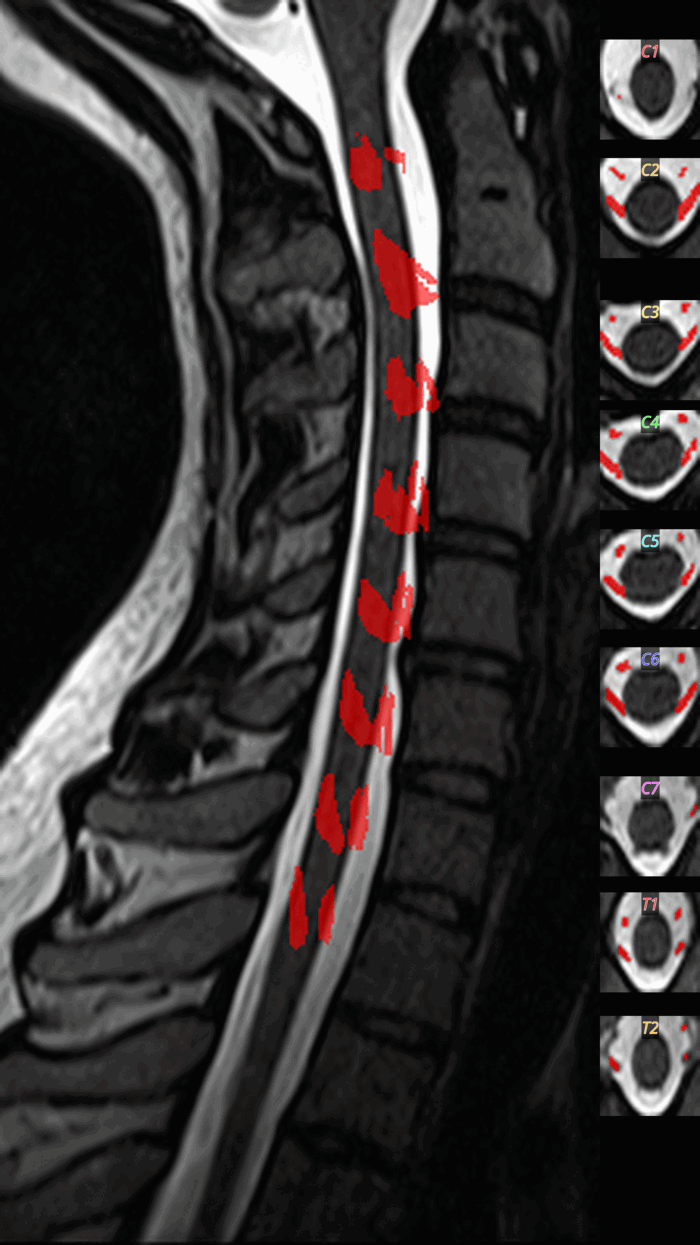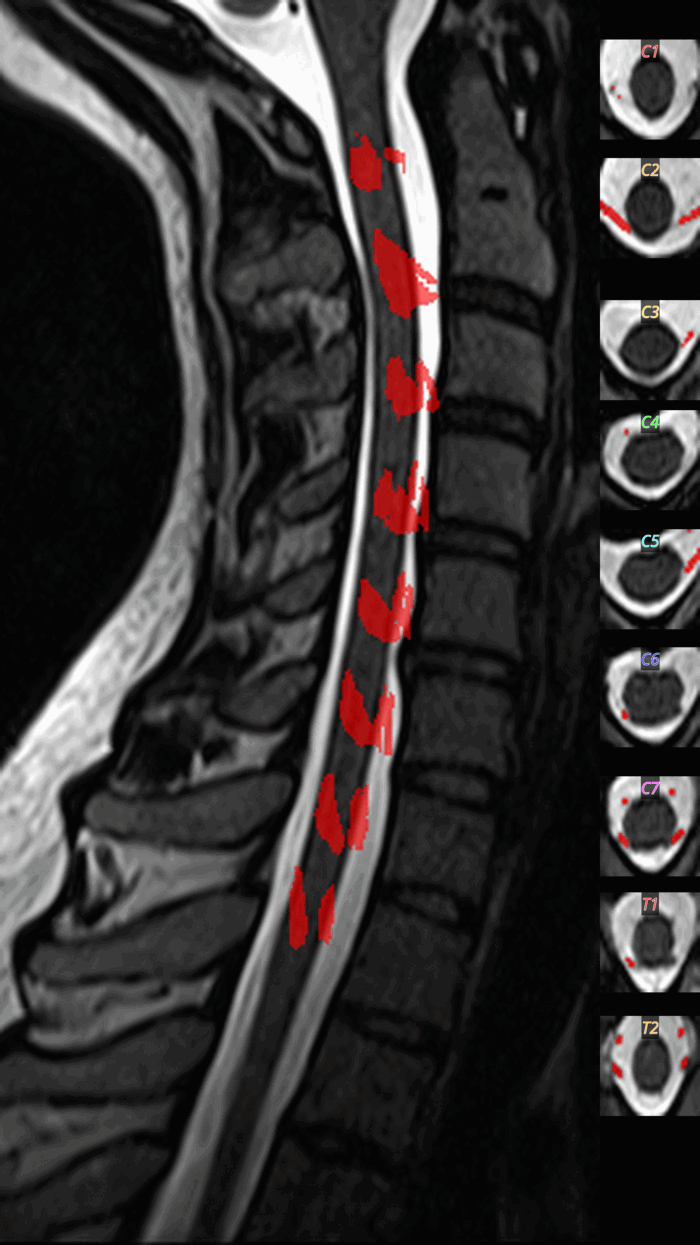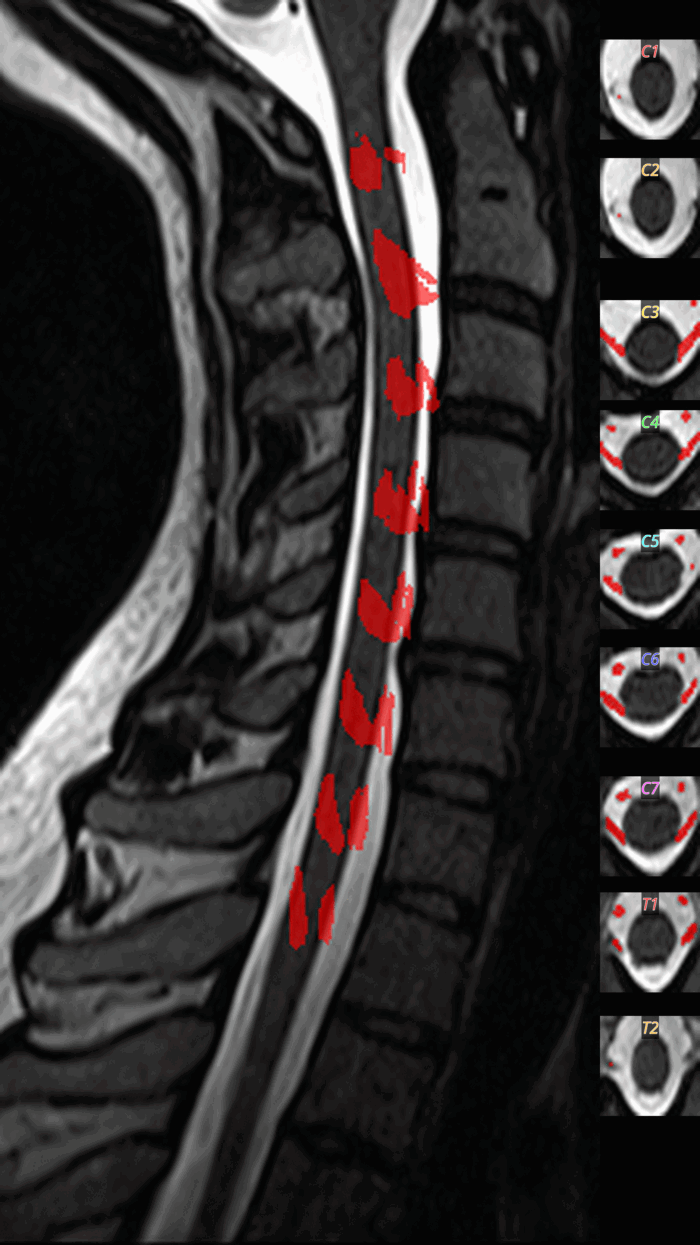
QC: Rootlets Montage
What to look for:
- ✅ Rootlets appear at dorsal-lateral positions
- ✅ Regular spacing along the cord
- ❌ FAIL: Rootlets detected ventrally (false positives)
S2.4: Template Registration
Strategy
Dual-path when rootlets available:
- Disc-based registration - vertebral disc labels as landmarks
- Rootlet-based registration - nerve rootlet entry points
Command: sct_register_to_template
Output Warps
*_from-cordref_to-PAM50_warp.nii.gz(forward)*_from-PAM50_to-cordref_warp.nii.gz(inverse)
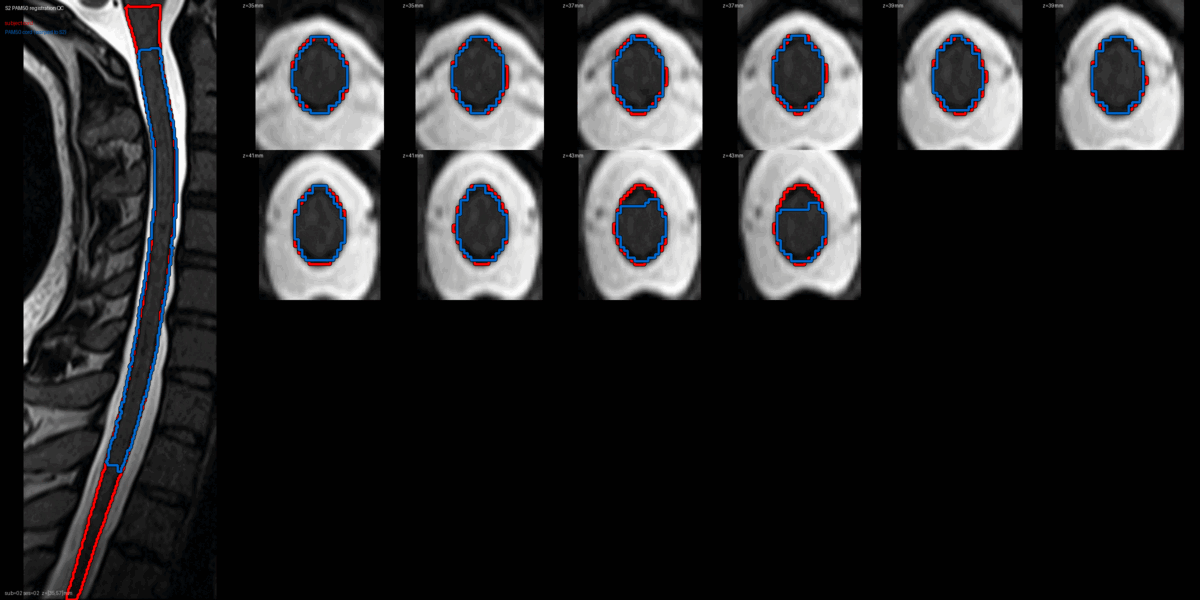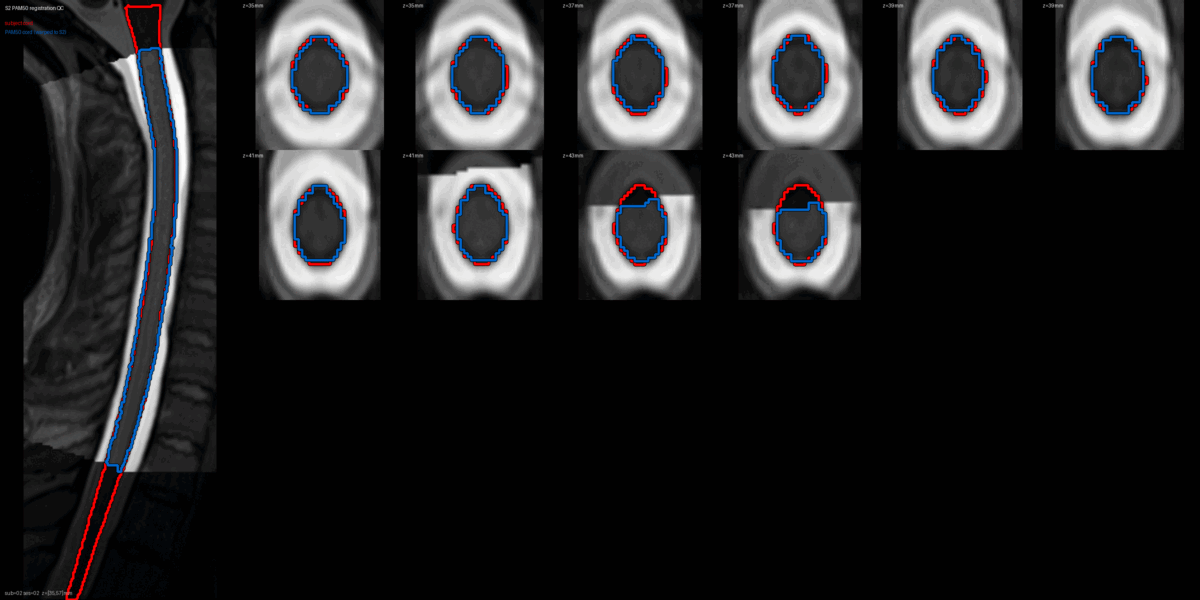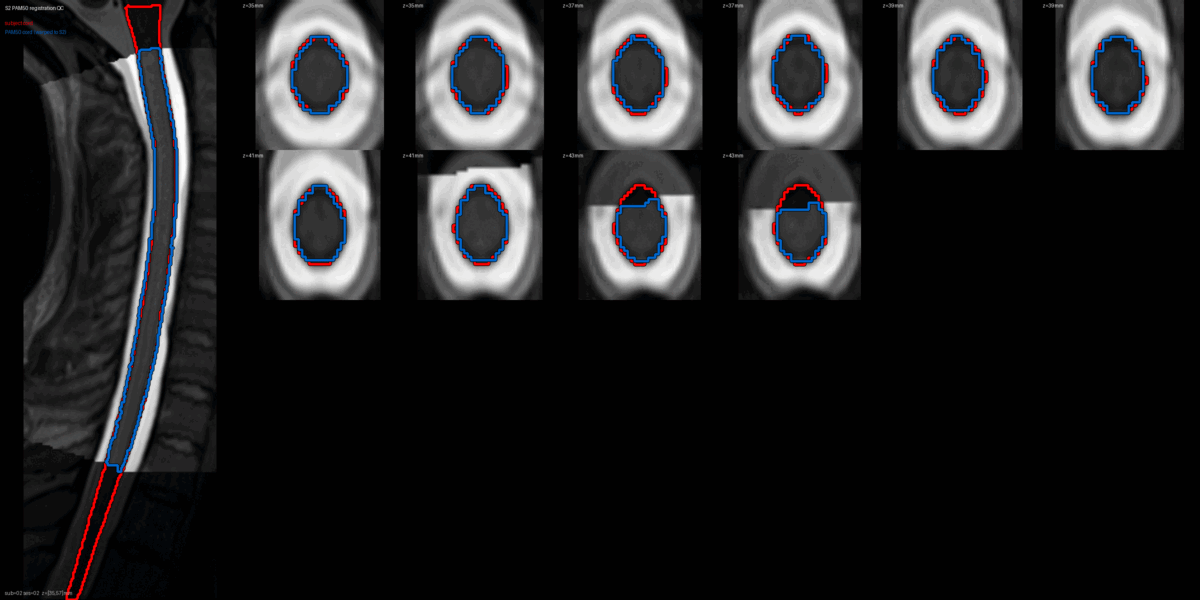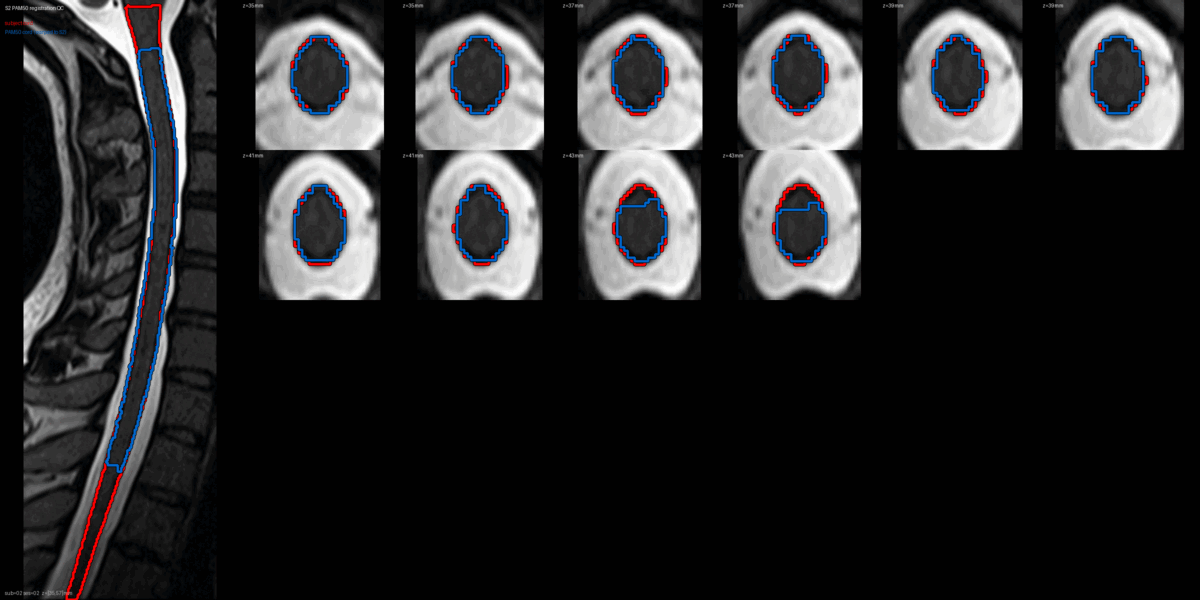
QC: PAM50 Registration Overlay
This animated GIF alternates between subject anatomy and template overlay.
What to look for:
- ✅ Cord contours align between subject and template
- ✅ No large rotational offsets
- ❌ FAIL: Significant misalignment (>5mm offset)
Outputs
Derivatives
derivatives/spinalfmriprep/{dataset}/sub-{id}/anat/
├── sub-{id}_desc-cordref_{T1w|T2w}.nii.gz
├── sub-{id}_desc-cord_dseg_{T1w|T2w}.nii.gz
├── sub-{id}_desc-canal_dseg_{T1w|T2w}.nii.gz
├── sub-{id}_desc-totalspineseg_dseg_{T1w|T2w}.nii.gz
├── sub-{id}_desc-vertebral_labels_{T1w|T2w}.nii.gz
├── sub-{id}_desc-disc_labels_{T1w|T2w}.nii.gz
└── sub-{id}_desc-rootlets_dseg_{T1w|T2w}.nii.gz (if eligible)
derivatives/spinalfmriprep/{dataset}/sub-{id}/xfm/
├── sub-{id}_from-cordref_to-PAM50_warp.nii.gz
└── sub-{id}_from-PAM50_to-cordref_warp.nii.gz
derivatives/spinalfmriprep/{dataset}/sub-{id}/figures/
├── sub-{id}_desc-S2_crop_box_sagittal.png
├── sub-{id}_desc-S2_cordmask_montage.png
├── sub-{id}_desc-S2_totalspineseg_montage.png
├── sub-{id}_desc-S2_rootlets_montage.gif (conditional)
└── sub-{id}_desc-S2_pam50_reg_overlay.gif
CLI Usage
# Run S2 for a single dataset
poetry run spinalfmriprep run S2_anat_cordref \
--dataset-key <KEY> \
--datasets-local config/datasets_local.yaml \
--out work/wf_001
# Run S2 for all regression datasets
poetry run spinalfmriprep run S2_anat_cordref \
--scope reg \
--datasets-local config/datasets_local.yaml \
--out work/wf_001
# Validate outputs
poetry run spinalfmriprep check S2_anat_cordref --out work/wf_001
References
- TotalSpineSeg: Löffler et al. (2024). GitHub
- SCT: De Leener et al. NeuroImage 145:24-43 (2017). DOI
- PAM50: De Leener et al. NeuroImage 165:170-179 (2018). DOI
Last updated: January 2026